Researchers use Anvil to create AI model for medical image diagnosis
Researchers from Arizona State University utilized the Anvil supercomputer to develop and deploy a fully open AI (artificial intelligence) foundation model for diagnosing diseases based on medical imaging. The new model, called Ark+, was recently published in the July 10 issue of Nature1. Ark+ was applied and thoroughly tested in the field of chest radiography. The researchers expect the underlying concept for Ark+ to be applicable across domains, including biology, chemistry, physics, and medicine, and hope that their work will inspire others to share code and datasets in order to accelerate open science and democratize AI.
Dr. Jianming Liang is a Professor in the College of Health Solutions at Arizona State University. Liang heads a research group at the university composed of himself and his graduate students. Together, they study annotation-efficient deep learning models for medical image analysis. Traditional deep learning models used for medical imaging analysis rely heavily on large, annotated datasets. However, annotating this data is a costly and inefficient process requiring qualified annotators to mark every image individually. Liang’s group seeks to address this challenge by exploring and developing deep learning models trained on datasets with limited annotated images. The research group has used Anvil for numerous experiments, but recently focused on developing Ark+, a fully open foundation model used in chest radiography.
Ark+ was designed to help democratize access to diagnostic capabilities. By creating an open AI model that can accurately assess medical images, Liang and his team can help support medical facilities and enable quicker, and potentially better, diagnoses, especially in communities that lack radiological expertise. A major aspect of Ark+ is that it is fully open. Many proprietary foundation models are not, making it difficult for researchers and developers to build on existing works, improve the model, or tailor it to their specific needs. The vision behind Ark+ was to create a powerful, robust foundation model that could be trained by aggregating public datasets while retaining the option to use federated private data. In this way, Ark+ remains fully accessible and usable to the public.
“AI and deep learning (DL) is revolutionizing many aspects of our lives, but the greatest impact of AI/DL has yet to come to healthcare via computer-aided diagnosis (CAD),” says Liang. “To build AI/DL-enabled CAD, we must first overcome a technological barrier: AI/DL requires massive amounts of carefully annotated data for training, but annotating medical data, especially medical images, is not only tedious, laborious, and time-consuming, but it also demands costly, specialty-oriented expertise.”
Liang continues, “Our research aims to address this annotation-dearth challenge in medical imaging by developing novel self- and full-supervised pretraining strategies, thereby relieving the annotation demand for training downstream (target) tasks. In the case of self-supervised pretraining, we utilize billions of image patches extracted from the original images for deep models to ‘understand’ anatomy autodidactically. In the case of full-supervised pretraining, we leverage any accessible, heterogeneous expert labels associated with any available datasets to ‘teach’ deep models to recognize disease patterns in images. Ark+ is a full-supervised pretraining strategy, representing a methodological breakthrough for learning one high-performance model from a multitude of datasets that are labeled differently.”
Ark+ was 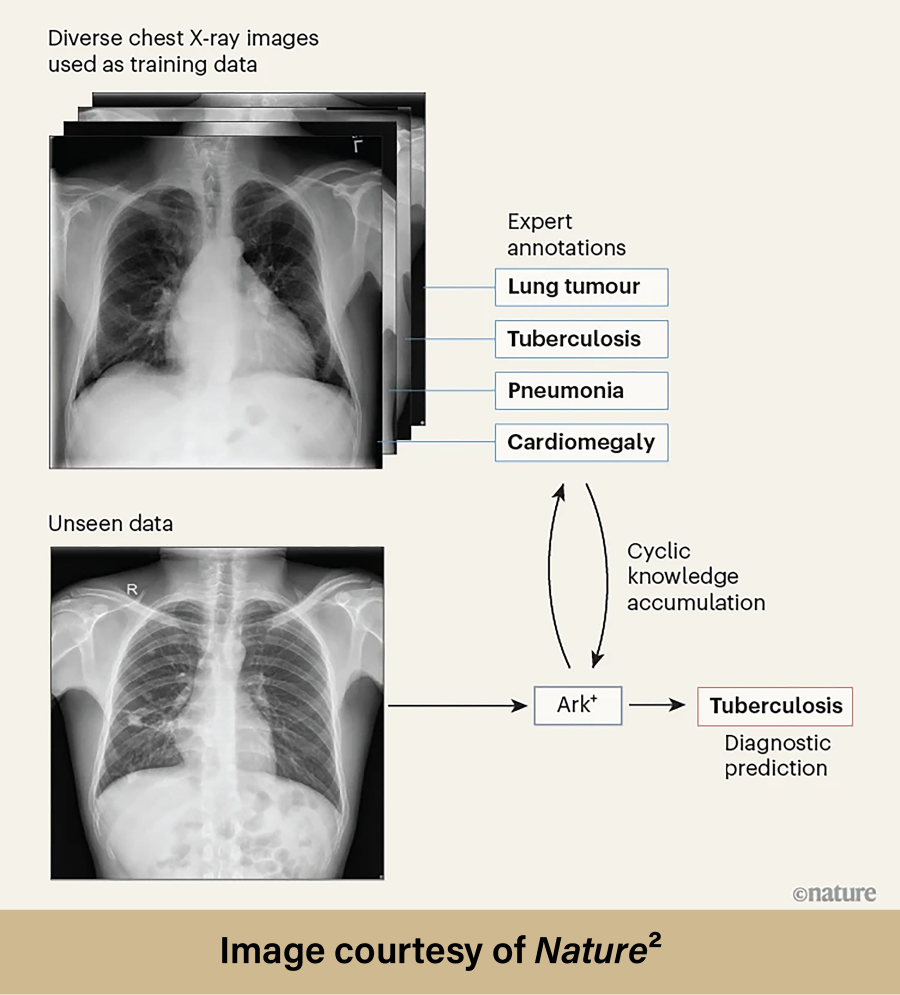 pretrained by cyclically accruing and reusing the knowledge contained within six public datasets. These six datasets contained over 700,000 chest radiography images which had already been annotated by experts. Once Ark+ was trained, Liang and the research group assessed its ability to properly diagnose thoracic diseases. Testing began by using data similar to the data the model was trained on, highlighting Ark+’s effectiveness when assessing images within familiar contexts. Testing then progressed to using “unseen” datasets; these images could be from different clinical settings or hospitals with different imaging protocols and varied patient populations. Without testing using “unseen” datasets, the researchers would have no way of knowing if Ark+ could perform in real-world scenarios that would likely differ drastically from the model’s training environment. Eight scenarios were used to evaluate Ark+ within each testing stage (which included a total of ten different datasets):
pretrained by cyclically accruing and reusing the knowledge contained within six public datasets. These six datasets contained over 700,000 chest radiography images which had already been annotated by experts. Once Ark+ was trained, Liang and the research group assessed its ability to properly diagnose thoracic diseases. Testing began by using data similar to the data the model was trained on, highlighting Ark+’s effectiveness when assessing images within familiar contexts. Testing then progressed to using “unseen” datasets; these images could be from different clinical settings or hospitals with different imaging protocols and varied patient populations. Without testing using “unseen” datasets, the researchers would have no way of knowing if Ark+ could perform in real-world scenarios that would likely differ drastically from the model’s training environment. Eight scenarios were used to evaluate Ark+ within each testing stage (which included a total of ten different datasets):
- Diagnosing common thoracic diseases
- Adapting to evolving diagnostic needs
- Learning to diagnose rare conditions from a few samples
- Handling long-tailed thoracic diseases
- Adjusting to shifts in the diagnostic setting without training
- Tolerating sex-related bias
- Responding to novel thoracic diseases
- Using private data while preserving patient privacy with federated pretraining
After extensively testing Ark+’s performance across these eight scenarios, the research group found the model to be more successful than anticipated. The results were overwhelmingly positive. Ark+ proved to be generalizable, adaptable, robust, and extensible while remaining open, public, light, and affordable. In direct comparison, Ark+ outperformed nine other foundation models. Liang’s research shows that accruing and reusing knowledge from numerous public datasets containing expert annotations can create a better AI model than proprietary ones trained on unusually large data. Of course, developing and testing Ark+ would have been impossible without the use of a high-performance computing (HPC) resource such as Anvil. AI models are dependent upon access to powerful GPUs, and Anvil provides these cutting-edge resources to AI researchers nationwide.
“Given the computational intensity nature of pretraining strategies, we rely on the powerful GPUs provided by the Anvil supercomputer. These advanced computing capabilities enable us to support our diverse range of computational and data-intensive research effectively. By leveraging these computing resources, our research could accelerate significantly, enabling deep learning algorithms to better generalize real-world clinical data. Ultimately, this advancement will enhance the effectiveness of computer-aided diagnosis at the point of care.”
To view the full development and testing procedures of Ark+, as well as the comprehensive results, please read the Liang research group’s Nature publication: A fully open AI foundation model applied to chest radiography
To learn more about High-Performance Computing and how it can help you, please visit our “Why HPC?” page.
Anvil is one of Purdue University’s most powerful supercomputers, providing researchers from diverse backgrounds with advanced computing capabilities. Built through a $10 million system acquisition grant from the National Science Foundation (NSF), Anvil supports scientific discovery by providing resources through the NSF’s Advanced Cyberinfrastructure Coordination Ecosystem: Services & Support (ACCESS), a program that serves tens of thousands of researchers across the United States.
Researchers may request access to Anvil via the ACCESS allocations process. More information about Anvil is available on Purdue’s Anvil website. Anyone with questions should contact anvil@purdue.edu. Anvil is funded under NSF award No. 2005632.
- Ma, D., Pang, J., Gotway, M.B. and Liang, J.. A fully open AI foundation model applied to chest radiography. Nature 643, 488–498 (2025). https://doi.org/10.1038/s41586-025-09079-8
- Kim N. An open AI model could help medical experts to interpret chest X-rays. Nature. Published online June 11, 2025. doi:https://doi.org/10.1038/d41586-025-01525-x
Written by: Jonathan Poole, poole43@purdue.edu
